
Research Details
-
Project Leaders
Prof. Dr. Heribert Schunkert
German Heart Centre Munich
Department of Cardiovascular Diseases, TUM
schunkert@dhm.mhn.de
Dr. Zhifen Chen (Principal Investigator)
chenz@dhm.mhn.de -
Research Staff
Dr. Ling Li (Postdoc)
lingjoyo.li@tum.de
Anastasia Diagel (PhD student)
anastasia.diagel@tum.de
Amos Romer (MD student)
amos.romer@tum.de
Xiaoning Song (MD student)
xiaoning.song@tum.de
Miaomiao Li (MD student)
miaomiao.li@tum.de
Constanze Lehertshuber (MD student)
c.lehertshuber@tum.de
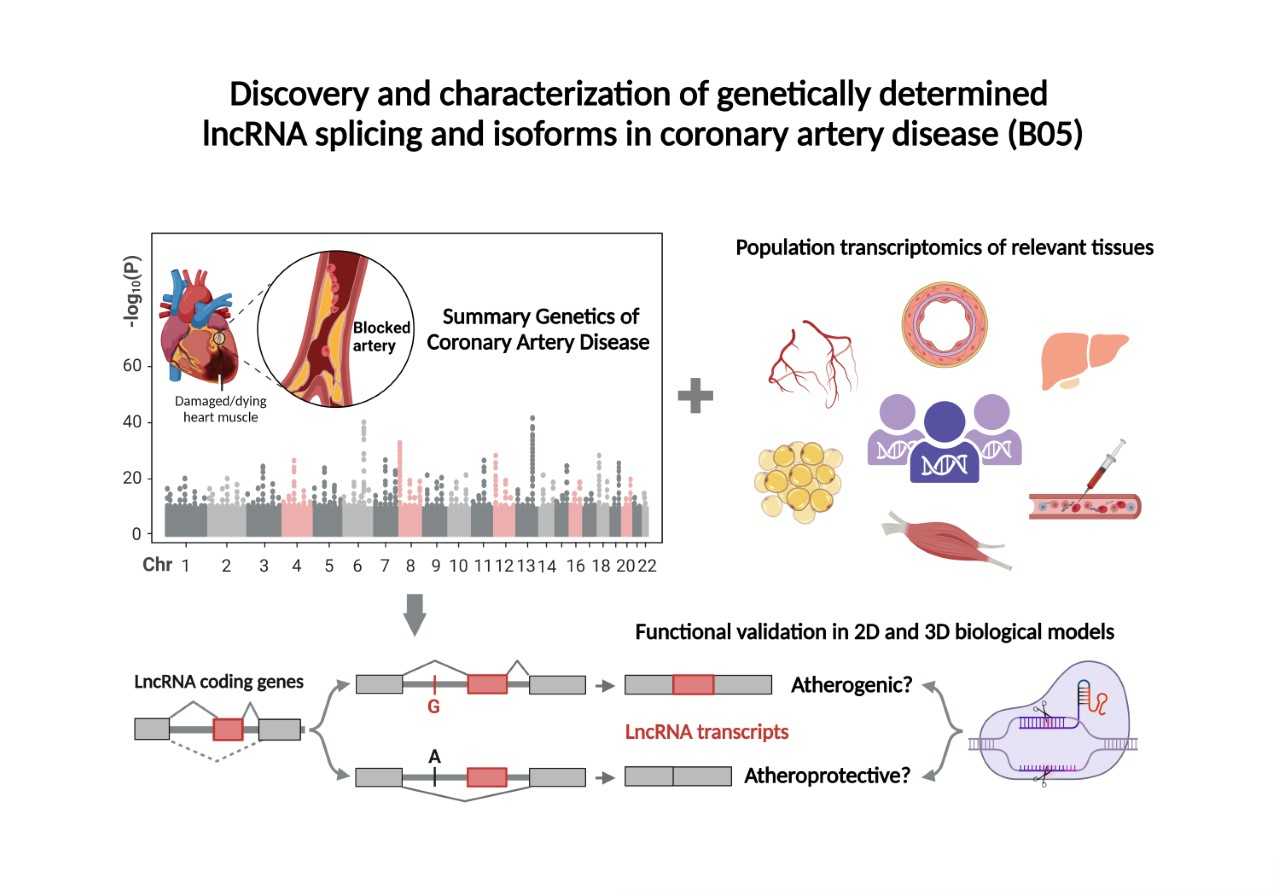
Despite the recent success of genetic studies in identifying chromosomal loci that associate with coronary artery disease (CAD), the roles of long noncoding RNAS (ncRNAs), their isoforms, and splice variants have been under-investigated. In the 1st funding period, we performed the first CAD transcriptome-wide association study (TWAS), and identified 6 lncRNAs, whose genetically regulated expression pattern was genome-wide significantly associated with CAD. We observed that genetic variants regulated distinct lncRNA isoforms and these isoforms might have differential effects on CAD risk. Two isoforms, CH-isoAS1 at CDH13 genetic locus and LINC00310-204 at LINC00310/KCNE2 locus were observed in human serum extracellular vesicles (EVs), which might mediate intercellular communications. Both genetic and functional data suggest that the CH-isoAS1 likely plays a causal role in mediating CAD risk and is cardioprotective. In this proposal, firstly, we will further confirm CH-isoAS1 mediated immune-vascular crosstalk and test its therapeutic value in an 3D engineered human artery model (eArtery). Secondly, we will focus on characterizing specific function(s) of LINC00310 isoforms in CAD and therapeutic value of the causal LINC00310 isoform(s) using advanced RNA-editing and -tracing tools including dCas9-based transcriptional activation and dCas13-medicated RNA monitoring in human pluripotent stem cell (hiPSC)-derived 2D and 3D models. Finally, we will perform a splicing wide association study (SWAS) to systematically identify splice variants and isoforms of lncRNAs in CAD using tissue transcriptomic data of ~3000 participants and genomic data of over one million individuals. Based on our findings, we hope to better understand the roles of lncRNA splicing and isoforms in CAD and prioritize novel diagnostic markers and therapeutic transcript targets for the disease.





